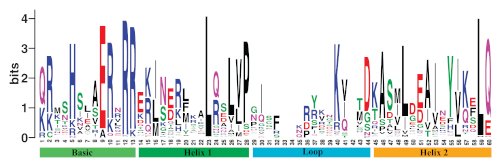
As it is known, the sequences of the bHLH transcription factor proteins have a certain structure like other protein sequences. bHLHs; It consists of 4 parts: Basic, Helix 1, Loop and Helix 2. All of these domains make up the DNA-binding region about 53 to 50 amino acids long. For example, 202 bHLH gene members, 60 to 742 amino acids long, belonging to the poplar plant were identified in a previous study. Logos are created to understand the structure of these bHLH sequences and to use them in various analyzes. HMMER's hmmbuild subtool, a Hidden Markov Model tool, can be used to create logos. A logo provides a compact graphical representation of an alignment, representing each column with a stack of letters. The total height of each stack corresponds to a measure of the invariance of the column – typically, it is the information content of that position. The height of each letter within a stack depends on the frequency of that letter at that position. Logos were originally devised to represent the extent of letter conservation in each column of an alignment, and were later generalized to show letter and gap probabilities of a profile HMM. These logos The following photo and the photo below show sample bHLH logos.
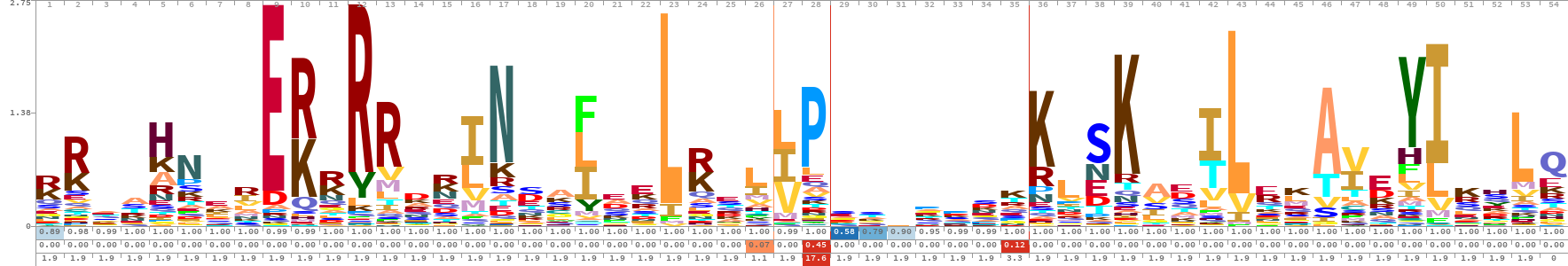
The first of the 3 rows filled with numbers in the photo above contains the occupancy value at each position, the second Insert Probability, and the third the Expected Insert Length value.
In addition, phylogenetic trees are frequently used in the literature, similar to these motifs. Thanks to phylogenetic trees, the functions and relationships of transcription factors can be studied more practically. Below is an example of the phlogenetic tree of bHLH proteins in the poplar plant.
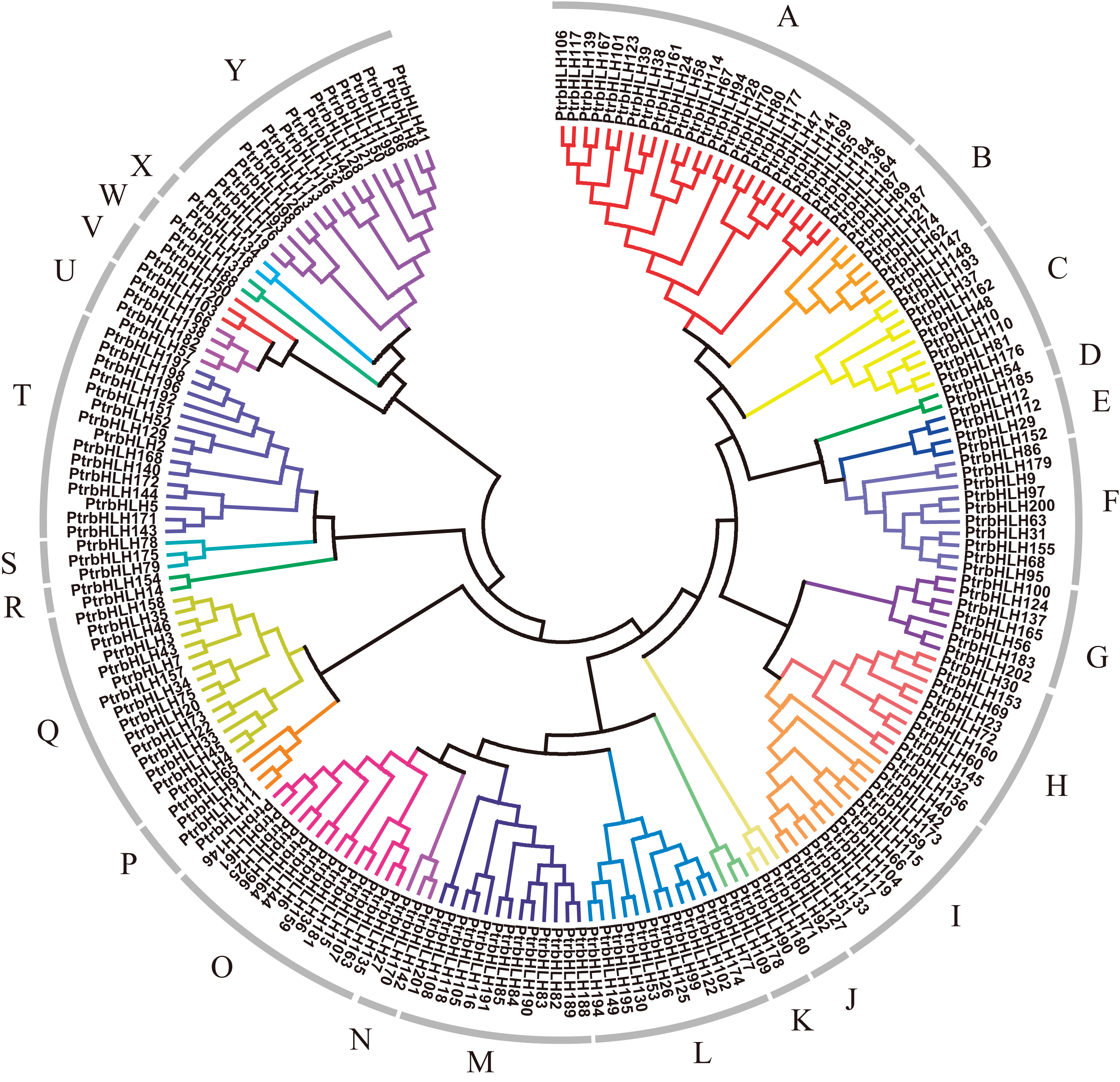
According to this phylogenetic tree, as expected, members in each group, especially closely related proteins, have the same or similar conserved motifs. Obtained 15 conserved protein sequence motifs based on the MEME. It is shown that motifs 1, 2, 3, 4, 6, 7 were annotated to the bHLH domain, by use of Pfam and InterProScan. Motifs 2 and 4 share the E-box/N-box specific site. Motif 5 is the ACT domain. Motif 8 is achaete-scute transcription factor-related. Motif 9 is the bHLH-MYC and R2R3-MYB transcription factors N-terminal. The others have no annotation.
Based on MEME, all the bHLH family genes contain the motif which was annotated as the bHLH domain, except bHLH202 without any motif. Group A, B, C, E mainly contain motifs 1 and 3; 13 groups largely share motif 2; group O harbors motif 4; 7 groups mainly have motifs 6 and 7; and several proteins in multiple groups contain motifs 1, 2, or 3. Regarding other motifs, motif 5 is mainly distributed in group K–R; motif 8 in group T; motifs 9, 11, 12, 13 in group Q; motif 10 in group B and Y; motif 14 in group I; motif 15 in group K, N, O, P, Q. In addition, group F only contains motif 2, group U only harbors motifs 6 and 7, and group Y only has motifs 2 and 10. Motif 4 only occurs in group O.
References
[1]: T. J. Wheeler, J. Clements and R. D. Finn, "Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models," BMC Bioinformatics, vol. 15, no. 7.
[2]: K. Zhao, S. Li, W. Yao, B. Zhou, T. Jiang, "Characterization of the basic helix–loop–helix gene family and its tissue-differential expression in response to salt stress in poplar; PeerJ, vol. 6.
[3]: https://pfam.xfam.org/family/PF00010#tabview=tab4